Introduction
Acute myeloid leukemia (AML) is a biologically highly heterogeneous disease. An increasing number of genetic abnormalities responsible for its pathogenesis are being incorporated in latest classifications to refine AML diagnosis. Myelodysplasia-related AML (AML-MR) is a broad category defined by mutations in a significant number of genes and cytogenetic alterations. Besides, there are still patients that must be classified according to phenotypic characteristics (AML defined by differentiation, AML-DD). Both categories still constitute heterogeneous groups in which further investigation is warranted to better define their underlying pathogenesis. Transcriptome sequencing has proven to be useful for fusion genes identification. In this study, we aimed to identify fusion genes by RNAseq in AML without recurrent genetic abnormalities and correlate them with clinical and phenotypic data.
Methods
One hundred and nine patients diagnosed with AML without recurrent genetic alterations between 1998 and 2020 in hospitals from CETLAM cooperative group were included. RNA samples collected at diagnosis were sequenced following a paired-end Illumina protocol. Five additional RNA samples from healthy subjects were sequenced for filtering purposes. Fusions were detected using STAR-Fusion version 1.9.0, against a reference based on GRCh38 and annotation gencode 35. In 66 cases, mutational information was available from whole-exome or targeted next-generation sequencing. In the other 43 cases, RNAseq data were used for variant calling in 40 recurrently mutated genes. Statistical analysis was done with R (version 4.04).
Results
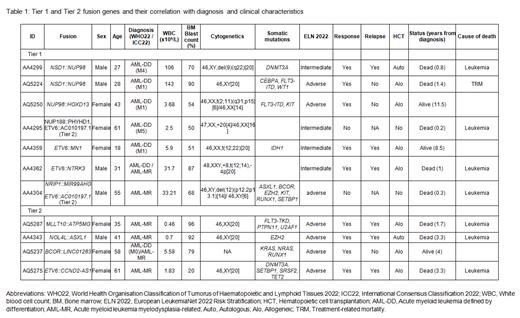
Fusion genes candidates were found in 53 of the 109 patients (49%). Likely artifacts were removed by ruling out fusions also found in healthy samples, fusions affecting immunoglobulin genes, fusions involving the same gene with multiple partners within the same sample or those found in more than one case without being a known fusion. After this process, 46 fusion candidates found in 29 different patients (27%) were manually classified as Tier 1 (well-known fusions, n=7), Tier 2 (fusions affecting recurrently mutated genes in AML without a functional validation, n=6), Tier 3 (fusions of unknown significance, n=25) and Tier 4 (fusions found in healthy tissues, n=8). Tier 1 and Tier 2 fusion genes are shown in Table 1. NSD1::NUP98 was the only fusion gene found twice, with the exact same breakpoint, in two male patients of young age (27 and 28 years). ETV6 was the gene more often involved in fusions (n=4), always with a different partner.
Patient characteristics of Tier 1 or Tier 2 fusion carriers (FC) are detailed in Table 1. Diagnosis in this group consisted of AML-DD (n=7) and AML-MR (n=4). FC were younger at diagnosis (41 years vs 58 years, P=0.005) and showed a higher median bone marrow blast count (70% vs 54%, P=0.07). Four of the eleven FC (36%) had a normal karyotype, in contrast to 58% in the whole cohort ( P=0.18). No difference was found either in white blood cell count, peripheral blast count or the presence of morphological dysplasia.
Ninety-three patients were eligible for intensive treatment. Two FC were primary refractory (75% vs 89%, P=0.18) and died without ever achieving a complete response (CR). Nine patients received transplantation in first CR (autologous, n=2; allogeneic, n=7), one of them dying of transplant-related complications. From the remaining six patients, four experienced relapse. Only one of these patients achieved a sustained remission after a second allogeneic transplantation. Two-year overall survival was similar between FC group and the rest of the cohort (46% vs 53%, P=0.6)
Of note, Tier 1 fusions NUP188::PHYHD1 and ETV6::NTRK3 were the only driver events found in two cases. The latter patient presented with a refractory relapse and died 11 months after diagnosis, though nowadays he could have benefit of NTRK inhibitors. In another case, the identification of MLLT10::ATP5MG was useful as molecular marker during follow-up.
Conclusions
Transcriptome sequencing by RNAseq identifies a considerable amount of fusion genes in AML. Ten percent of our cohort of patients with AML without recurrent genetic abnormalities have at least one Tier 1 or Tier 2 fusion. Infrequent fusions may be driver events in AML with clinical implications in terms of diagnosis, prognosis and targeted therapy.
Disclosures
Jimenez-Vicente:Pfizer: Other: Travel Grants; Abbvie: Other: Speaker, Travel Grants. Díaz-Beyá:Bristol Myers Squibb: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Astellas: Consultancy, Honoraria; Jazz Pharma: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; Novartis: Consultancy, Honoraria. Esteve:Pfizer: Research Funding; Kronos Bio: Research Funding; Gilead: Consultancy; Astellas: Consultancy; Jazz Pharmaceuticals: Consultancy, Research Funding; Abbvie: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal